Billions of years before humans developed hard drives, evolution chose DNA to store its most precious information: the genetic code. Over time DNA became so proficient at this task that every known life-form on earth uses it. With recent technological breakthroughs that allow us to easily “read” and “write” DNA, scientists are now repurposing this age-old molecule to store new types of information—the kind that humans are generating at an exponential rate in the age of big data.
The concept of repurposing DNA to store information beyond genetic code has been discussed extensively. After all, the 1s and 0s of computer code are bumping up against the limits of physics. One of the challenges to safely storing all the data we create was exposed recently, when Myspace—once the most popular social network—announced that a decade’s worth of data may have been irreparably lost in a server-migration project. The long-term protection of data, like those of a Web site that rebooted after a period of dormancy, exposes where existing technologies are vulnerable and clunky. And it’s not just a spatial problem: significant energy is needed to maintain data storage.
The properties of DNA have the potential to get around these issues. For one thing, DNA’s double-helix structure is perfectly suited for information storage because knowing the sequence of one strand automatically tells you the sequence of the other strand. DNA is also stable for extended periods, which means the integrity and accuracy of information can be maintained. For example, in 2017 scientists analyzed DNA isolated from human remains that were 8,100 years old. These remains were not even stored in ideal conditions the entire time. If kept in a cool, dry environment, DNA can almost certainly last tens of thousands of years. DNA is also stable for long stretches, which means the integrity and accuracy of information can be maintained.
On supporting science journalism
If you're enjoying this article, consider supporting our award-winning journalism by subscribing. By purchasing a subscription you are helping to ensure the future of impactful stories about the discoveries and ideas shaping our world today.
Perhaps the most compelling aspect of the double helix, however, is that it can fold into an extraordinarily dense structure. For comparison, every individual human cell contains a nucleus with a diameter of approximately 0.00001 meter. Yet if the DNA inside a single nucleus was stretched out, it would reach two meters. Put another way, if the DNA in a person was strung together, it would extend 100 trillion meters. In 2014 scientists calculated that it is theoretically possible to store 455 exabytes of data in a single gram of DNA. This information-storage density is about a millionfold higher than the physical storage density in hard drives.
Although DNA has commonly been thought of as a storage medium, there are still significant scientific, economic and ethical hurdles to overcome before it might replace traditional hard drives. In the meantime, DNA is becoming more widely—and immediately—useful as a broader form of information technology. DNA has been used, for instance, to record old Hollywood films, preserving the classics in genetic code instead of fragile microfilm. Even more recently, DNA has been used as a tool to design safer gene therapies, speed up anticancer drug development and even generate what is perhaps the first genetic “live stream” of a living organism. On the frontiers of this evolving field, DNA is being pursued not just for long-term data storage but for facilitation of data generation at unprecedented speed. That is because DNA is more scalable than any other molecule in both directions: it allows us to dramatically expand the amount of data we create and shrink the resources needed to store them.
Accelerating New Nanoparticles
In recent years scientists have increasingly used DNA as a molecular recorder to understand and keep track of their experimental results. In many cases, this process involves DNA bar coding: To label and track the result of an individual experiment, scientists use a known DNA sequence to serve as a molecular tag. For example, one experimental outcome might be associated with the DNA sequence ACTATC, whereas another outcome might be associated with a TCTGAT, and so on.
DNA bar coding has been around since the early 1990s, when Richard Lerner and the late Sydney Brenner, both then at the Scripps Research Institute, proposed it as a way to track chemical reactions. Their concept was tremendously innovative but ahead of its time: technologies that easily and inexpensively read out DNA had not yet been developed. Its potential was only realized after many scientists made contributions to nucleotide chemistry, microfluidics and other approaches, which together enabled the advent of what is called next-generation sequencing. A major breakthrough came in 2005, when researchers reported that 25 million DNA bases were analyzed in a four-hour experiment.
Next-generation sequencing has continued to rapidly improve; it is now easy to read millions of DNA sequences at the same time, which means that thousands of experiments can be performed and analyzed simultaneously. Analyzing DNA bar code experiments with next-generation sequencing is its own form of data management: instead of testing ideas one at a time, scientists can make 20,000 predictions and test them all to see which is correct.
Biologists were the first to utilize DNA bar coding extensively. As it has become more accessible, researchers in many different fields, including chemical engineering and materials science, are using the technology to perform experiments at entirely new scales. In my laboratory at the Georgia Institute of Technology, for instance, engineers are using DNA bar codes to improve the design and function of nanoparticles so that they can safely deliver drugs to diseased cells. Nanotechnology, which relies primarily on physics and chemical engineering, may seem completely unrelated to DNA. But when you think of DNA as a way to track and store any data, its utility as an organizational tool becomes apparent.
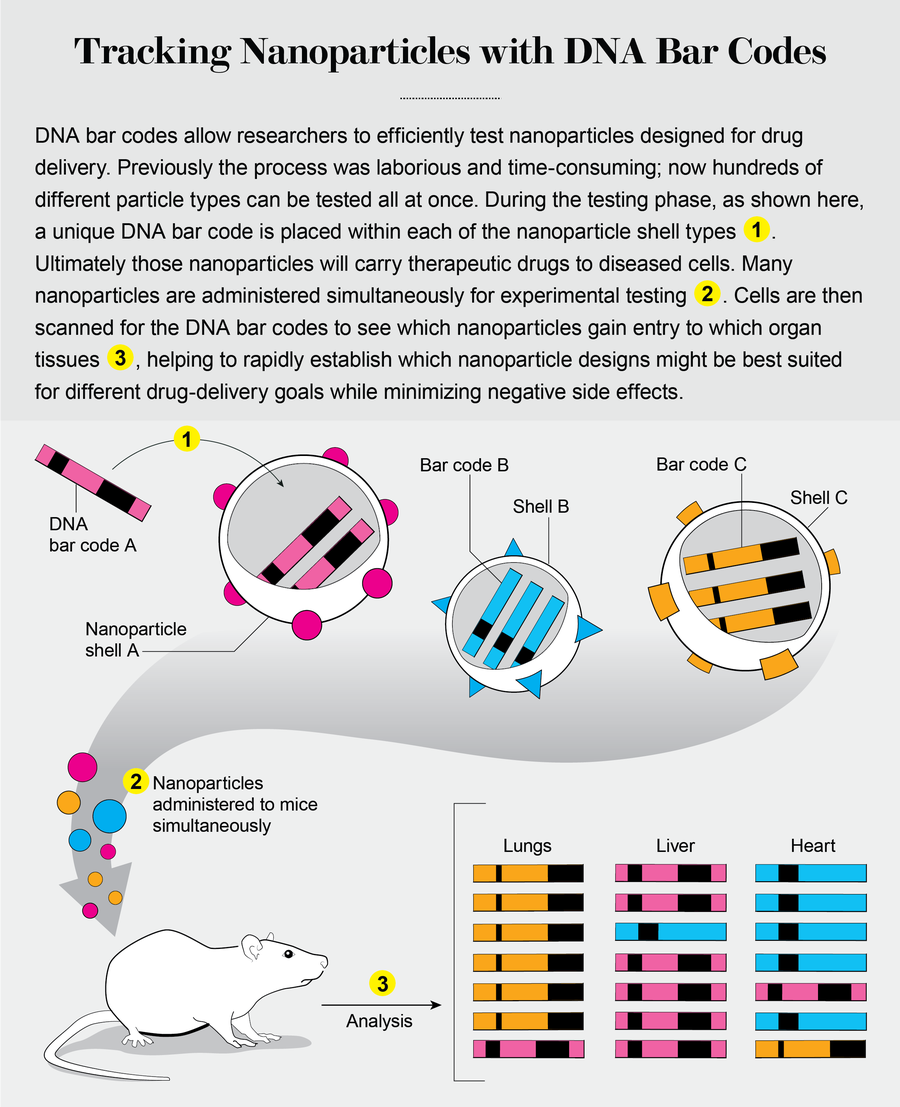
Credit: Jen Christiansen; Source: “A Direct Comparison of In Vitro and In Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation,” by Kalina Paunovska et al., in Nano Letters, Vol. 18, No. 3; March 14, 2018
One fundamental problem for nanotechnologists is that designing experiments to search for effective therapies is still far easier than performing them and analyzing the results. That is because the shape, size, charge, chemical composition and many other variables of individual nanoparticles can alter how well they deliver their genetic drugs to diseased cells. Additionally, these factors all interact with one another, making it a struggle for researchers to predict which nanoparticle will deliver its drug in the most targeted way. An obvious solution is to evaluate every nanoparticle one by one. But data from established pharmaceutical companies that have developed nanoparticles for RNA drugs have demonstrated that this type of testing can require several hundred million dollars to pull off.
That is where the storage capabilities of DNA can make big strides. To increase the number of nanoparticles we are able to test, we can design thousands of them with diverse chemical structures—large, positively charged spheres or small, neutrally charged triangles, for example—and assign each a DNA bar code.
Nanoparticle one, with chemical structure one, carries DNA bar code one. Nanoparticle two, with chemical structure two, carries DNA bar code two. We repeat this bar-coding process many times, thereby creating many different nanoparticles, each with its own unique molecular DNA tag. We can then administer hundreds of these nanoparticles to diseased cells. To identify the nanoparticle that most successfully delivered the drug, we use DNA sequencing to quantify the bar codes inside the cells.
The scale of such experiments is entirely new to nanomedicine. A “traditional” experiment in my field generates between one and five data points. By the end of 2019 my lab hopes to quantify how 500 different nanoparticles deliver gene therapies to 40 different cell types. Doing so is equivalent to running 20,000 experiments simultaneously.
As a result, we also needed to create a data-analysis pipeline capable of monitoring data quality, as well as helping us statistically test our results. First, we measured how well results from one replicated experiment predicted delivery in another. Once we knew the large data sets were reliable, we used statistics to ask whether certain nanoparticle traits—such as their size—affected delivery to target tissues. We found that the chemistry of the nanoparticle, not its size, dictated nanoparticle delivery. Using this approach, we hope to discover safe gene therapies more quickly, using far fewer resources. One of our goals is to identify a nanoparticle that can specifically deliver gene therapies that help kill tumors, thereby reducing side effects such as nausea and hair loss that accompany existing treatments.
We have already had some success. In 2018, by using very large data sets generated by DNA bar-coding experiments, we rapidly identified new nanoparticles that deliver gene therapies to endothelial cells, which line blood vessels, as well as several types of immune cells, which govern how our bodies respond to disease. This finding could change treatment by allowing us to change the activity of proteins in immune cells that are currently “undruggable,” meaning the proteins are hard to target with small-molecule drugs or antibodies. As a result of data published in journals that included the Proceedings of the National Academy of Sciences USA, Advanced Materials and the Journal of the American Chemical Society in 2018 and 2019, we received a flood of interest from other gene therapists and were able to start GuideRx, a bar-coding company that focuses on efficiently developing safe gene therapies.
DNA bar coding has now become so commonplace that it is being applied in different ways even within a single field. One example is cancer biology, which looks at how genetic mutations cause cancer and how new drugs can treat it. Drug resistance remains a major challenge in this field: patients often initially respond to a drug but relapse as it loses the ability to kill tumor cells.
Scientists in the lab of Todd Golub at Harvard University have used DNA bar coding to study such resistance. In 2016 they described how they used a virus to permanently insert a DNA bar code directly into the genome of cancer cells. Cancer cell type A received bar code sequence A; cancer cell type B received bar code B, and so on. The scientists mixed the different cells together, plated them on a dish and treated them with a cancer drug.
If the drug killed the cancer cell or slowed its growth, then the cell would not divide. But if the cell became resistant to the drug, then it divided rapidly. Thus, over time the relative amount of bar code sequence A increased if cell type A became resistant to the drug or, alternatively, decreased if cell type A was killed by the drug. By sequencing all the bar codes from surviving cells over time, the lab quantified how well all the cell types responded to the drug simultaneously.
Later that year the lab of Monte Winslow at Stanford University used DNA-bar-coded pancreatic cell lines to identify drugs that prevented the spread of cancer, or metastasis. The lab bar coded each cell line using a virus, then plated each cell line in its own well. Each well was then treated with an anticancer drug. In this way, drug one became associated with bar code one. Immediately thereafter, the scientists injected the cells into the bloodstream, and they later measured which cells spread to the lungs. By identifying the bar codes that were abundant or absent, the researchers identified drugs that respectively promoted or prevented metastasis.
In a third example, scientists at the Broad Institute of the Massachusetts Institute of Technology and Harvard University used DNA bar coding to study how all the genes in the genome affect a single cancer. The researchers first grew a very large number of cells and plated them in a large dish together. Then they used a gene-editing system to inactivate or, alternatively, activate all the genes in the genome one by one. The sequence of the gene whose expression had been modulated acted as the bar code. By treating the cells with a cancer drug and sequencing the DNA over time, the scientists could understand how every gene in the genome affects drug resistance.
In these approaches, DNA is acting both as a data-generating molecule, because it is required to perform all the experiments simultaneously, and as a data-storage molecule, because next-generation sequencing is used to analyze the DNA bar codes. The implications are stunning: the same techniques can be applied to autoimmune and neurological diseases and cardiovascular dysfunction. The full power of using DNA bar coding can be understood with a simple exercise. In the examples discussed earlier, replace the word “cancer” with a different disease or the word “resistance” with any desired drug response. In this way, DNA bar coding is positioned to fundamentally streamline early-stage drug development, thereby accelerating the path to effective therapies.
Reading vs. Writing
DNA bar coding relies on “reading” known DNA sequences. Until recently, however, it was not practically possible to “write” DNA sequences. Broadly speaking, I think of writing DNA as purposefully converting other forms of information—such as pictures, movies or biological states—into sequences that can be stored and read out later. Many of these new writing technologies are driven by gene-editing systems derived from clustered regularly interspaced short palindromic repeats (CRISPR). With rationally engineered CRISPR systems, scientists can write DNA sequences.
Several of the most recent advances exploit the way CRISPR systems naturally evolved to defend bacteria against viral attacks. More specifically, viruses attack bacteria by binding onto the bacterial surface, then inserting their viral DNA or RNA. To “remember” the virus for future attacks, bacteria evolved CRISPR systems that identify viral DNA or RNA and then insert small snippets of the DNA into their own genome. In other words, the bacteria are “writing,” or “recording,” a history of the viruses that have attacked them to defend themselves.

Double-helix structure of DNA makes for an ideal storage medium. But it is not yet able to replace traditional hard drives. Credit: Getty Images
By exploiting this mechanism, Seth Shipman, working in the lab of Harvard geneticist George Church and now at the University of California, San Francisco, used CRISPR to record images of a human hand directly into the genome of Escherichia coli. To accomplish this task, Shipman and his colleagues first expressed two proteins: Cas1 and Cas2. Together these proteins can acquire DNA nucleotides and insert them into the genome. The researchers then “fed” E. coli DNA sequences that encoded for pixels that—when sequenced together—created the image of a hand. Doing so required the scientists to assign different aspects of information to DNA. For example, in one case, A, C, G and T each stood for a different pixel color, whereas an associated DNA bar code sequence encoded the spatial position of the pixel within the entire image.
By sequencing the DNA from the E. coli, the authors then recapitulated the original image with more than 90 percent accuracy. Next, they repeated the experiment but with an important twist: they added the DNA at different times and included a method to analyze the position of the recorded DNA sequences, relative to one another. By measuring whether the sequences were added into the E. coli genome earlier or later, they were able to create a series of images, thereby encoding a movie. The researchers recorded a GIF from a part of the first motion picture, which was created by Eadweard Muybridge in 1878 and depicted a galloping horse. In a 2017 paper, they showed that they had reconstituted Muybridge’s famous movie by sequencing the bacterial genome.
Even more recently, scientists in the lab of Randall Platt at the Swiss Federal Institute of Technology Zurich (ETH Zurich) made a critical discovery that takes these approaches even further by targeting mRNA, which is a key molecular cousin of DNA. Instead of recording images encoded by unnatural DNA sequences, they used a CRISPR system from a different bacterial species to generate so-called living records of natural mRNA gene expression in bacteria. The combination of all the different mRNAs in a cell dictates which proteins are made and therefore all cellular function.
To record mRNA produced by a cell at different time points, scientists at Platt’s lab first screened CRISPR-Cas proteins derived from many different bacterial strains. This process allowed them to identify proteins capable of converting natural mRNA into DNA and encoding it into the genome. They found that Cas1 and Cas2 proteins from the bacterium Fusicatenibacter saccharivorans were capable of doing so. Through a series of elegant studies using specialized viruses, the team demonstrated in 2018 that the cells accurately recorded whether they had been previously exposed to oxidative stress, acidic conditions or even an herbicide.
These results were extremely exciting because they demonstrated that the genes naturally expressed by a cell at a given time could be recorded into the genome for later analysis. As Platt’s lab continues to improve this technology, it is increasingly feasible that cellular recording could become commonplace. This development would enable scientists to track how a cell has become cancerous, responds to infection over time and even ages.
The Ubiquity of DNA Storage
As DNA is used to generate, track and store information in an increasing number of fields, the most obvious question is whether DNA will eventually compete with conventional electronic storage devices to maintain all the digital data humans generate. Currently the answer is no—hard drives and flash memory devices are far better at keeping information than even the most advanced DNA systems.
But like all technologies, conventional electronic devices have limitations. They take up physical space and require specific environmental conditions; even the most durable ones are unlikely to survive more than a few decades. Given these issues, it may soon become hard to maintain all the data we are generating today.
DNA, by comparison, could almost certainly last tens of thousands of years if kept in cool, dry conditions. It is already routinely stored at −20 or even −80 degrees Celsius in labs that require very cold conditions and can also be stored in the kind of extreme heat that typical electronics cannot withstand. In 2015 Robert Grass and Wendelin Stark, both at ETH Zurich, showed that DNA stored in silica could withstand 70 degree C temperatures for a week without introducing any errors. And although hard drives can fit as much as one terabit per square inch, recent estimations suggest that all the information generated in the entire world could theoretically be held in less than a kilogram of DNA.
There are still significant technological advances that need to be overcome for DNA storage to become commonplace. The primary limitation is that storing information is not identical to extracting it. Getting data from a hard drive is nearly instantaneous; extracting them from DNA requires sequencing, which currently takes a few minutes to a day to complete. And despite huge leaps in DNA sequencers over the past few years, they remain large and expensive as compared with hard drives.
These barriers are not the only considerations we must tackle before DNA storage can reach its full potential. As a society, we need to acknowledge that the ubiquity of DNA sequencing will also mean that it will become even easier to track people while generating new vulnerabilities for data security. Examples of privacy concerns abound, both in the U.S. and globally.
DNA sequencing is already being used by police departments across the U.S. with little oversight. By asking people who are under arrest—even for minor crimes—for their DNA, the police are establishing large data banks of genetic information. Some have argued this is the 21st-century equivalent of old-fashioned fingerprinting, but there is a critical difference. Fingerprints identify a single individual; if one of your relatives provides his or her DNA, that person is releasing information that can identify you or anyone else in your family. In China, under the guise of a health program, officials have gathered genetic information from nearly 36 million people. This population includes many Uighurs—members of a Muslim ethnic group that experiences discrimination. It remains unclear how these data will be used by the government.
Currently these concerns around DNA storage involve a person’s genetic code itself—the discussion has been around protecting identity. But in the future, if other categories of information such as health care data, legal contracts and individual digital histories were stored in DNA, this scenario would launch even more questions about the vulnerability of DNA storage in the realms of both physical security and cybersecurity. Because so much information can be held in such a tiny space, how will data be distributed to avoid too much concentration in a single place? And even if extraction can be streamlined, how will data be routinely accessed and returned without exposing them to malicious hacks or accidental loss?
When I consider all the hard work—both scientific and ethical—that needs to be accomplished, it can seem daunting. I like to think about the Wright brothers because I grew up in the same Ohio town they did. Their first flight lasted 12 seconds and 37 meters. Sixty-six years later, without the advantages of modern computing, humans landed on the moon. These feats make me optimistic that we can harness the natural power of DNA over the next few decades and, by actively acknowledging its capability to do harm, help to ensure it mostly does good.

